Deep learning: new computational modelling techniques for genomics
As a data-driven science, genomics largely utilizes machine learning to capture dependencies in data and derive novel biological hypotheses. However, the ability to extract new insights from the exponentially increasing volume of genomics data requires more expressive machine learning models. By effectively leveraging large data sets, deep learning has transformed fields such as computer vision and natural language processing. Now, it is becoming the method of choice for many genomics modelling tasks, including predicting the impact of genetic variation on gene regulatory mechanisms such as DNA accessibility and splicing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
206,07 € per year
only 17,17 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
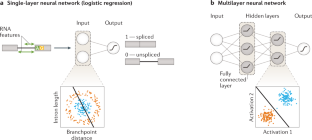
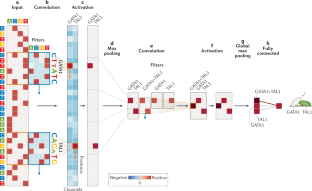
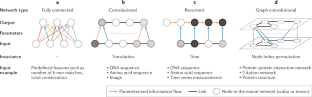
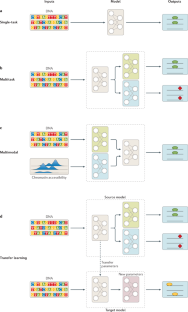
Current progress and open challenges for applying deep learning across the biosciences
Article Open access 01 April 2022
Predictive analyses of regulatory sequences with EUGENe
Article Open access 16 November 2023
A self-supervised deep learning method for data-efficient training in genomics
Article Open access 11 September 2023
References
- Hieter, P. & Boguski, M. Functional genomics: it’s all how you read it. Science278, 601–602 (1997). CASPubMedGoogle Scholar
- Brown, P. O. & Botstein, D. Exploring the new world of the genome with DNA microarrays. Nat. Genet.21, 33–37 (1999). CASPubMedGoogle Scholar
- Ozaki, K. et al. Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat. Genet.32, 650–654 (2002). CASPubMedGoogle Scholar
- Golub, T. R. et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science286, 531–537 (1999). CASPubMedGoogle Scholar
- Oliver, S. Guilt-by-association goes global. Nature403, 601–603 (2000). CASPubMedGoogle Scholar
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). PubMed CentralGoogle Scholar
- Murphy, K. P. Machine Learning: A Probabilistic Perspective (MIT Press, 2012).
- Bishop, C. M. Pattern Recognition and Machine Learning (Springer, New York, 2016).
- Libbrecht, M. W. & Noble, W. S. Machine learning applications in genetics and genomics. Nat. Rev. Genet.16, 321–332 (2015). CASPubMed CentralPubMedGoogle Scholar
- Durbin, R., Eddy, S. R., Krogh, A. & Mitchison, G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids (Cambridge Univ. Press, 1998).
- Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT Press, 2016). This textbook covers theoretical and practical aspects of deep learning with introductory sections on linear algebra and machine learning.
- Shi, S., Wang, Q., Xu, P. & Chu, X. in 2016 7th International Conference on Cloud Computing and Big Data (CCBD) 99–104 (IEEE, 2016).
- Krizhevsky, A., Sutskever, I. & Hinton, G. E. in Advances in Neural Information Processing Systems 25 (NIPS 2012) (eds Pereira, F., Burges, C. J. C., Bottou, L. & Weinberger, K. Q.) 1097–1105 (Curran Associates, Inc., 2012).
- Girshick, R., Donahue, J., Darrell, T. & Malik, J. in 2014 IEEE Conference on Computer Vision and Pattern Recognition 580–587 (IEEE, 2014).
- Long, J., Shelhamer, E. & Darrell, T. in 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 3431–3440 (IEEE, 2015).
- Hannun, A. et al. Deep speech: scaling up end-to-end speech recognition. Preprint at arXivhttps://arxiv.org/abs/1412.5567 (2014).
- Wu, Y. et al. Google’s neural machine translation system: bridging the gap between human and machine translation. Preprint at arXivhttps://arxiv.org/abs/1609.08144 (2016).
- Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol.33, 831–838 (2015). This paper describes a pioneering convolutional neural network application in genomics. CASPubMedGoogle Scholar
- Zhou, J. & Troyanskaya, O. G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods12, 931–934 (2015). This paper applies deep CNNs to predict chromatin features and transcription factor binding from DNA sequence and demonstrates its utility in non-coding variant effect prediction. CASPubMed CentralPubMedGoogle Scholar
- Zou, J. et al. A primer on deep learning in genomics. Nat. Genet.51, 12–18 (2019). CASPubMedGoogle Scholar
- Angermueller, C., Pärnamaa, T., Parts, L. & Stegle, O. Deep learning for computational biology. Mol. Syst. Biol.12, 878 (2016). PubMed CentralPubMedGoogle Scholar
- Min, S., Lee, B. & Yoon, S. Deep learning in bioinformatics. Brief. Bioinform.18, 851–869 (2017). PubMedGoogle Scholar
- Jones, W., Alasoo, K., Fishman, D. & Parts, L. Computational biology: deep learning. Emerg. Top. Life Sci.1, 257–274 (2017). PubMedGoogle Scholar
- Wainberg, M., Merico, D., Delong, A. & Frey, B. J. Deep learning in biomedicine. Nat. Biotechnol.36, 829–838 (2018). CASPubMedGoogle Scholar
- Ching, T. et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface15, 20170387 (2018). PubMed CentralPubMedGoogle Scholar
- Morgan, J. N. & Sonquist, J. A. Problems in the analysis of survey data, and a proposal. J. Am. Stat. Assoc.58, 415–434 (1963). Google Scholar
- Boser, B. E., Guyon, I. M. & Vapnik, V. N. A. in Proceedings of the Fifth Annual Workshop on Computational Learning Theory 144–152 (ACM, 1992).
- Breiman, L. Random forests. Mach. Learn.45, 5–32 (2001). Google Scholar
- Friedman, J. H. Greedy function approximation: a gradient boosting machine. Ann. Stat.29, 1189–1232 (2001). Google Scholar
- Xiong, H. Y. et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science347, 1254806 (2015). PubMedGoogle Scholar
- Jha, A., Gazzara, M. R. & Barash, Y. Integrative deep models for alternative splicing. Bioinformatics33, i274–i282 (2017). CASPubMed CentralPubMedGoogle Scholar
- Quang, D., Chen, Y. & Xie, X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics31, 761–763 (2015). CASPubMedGoogle Scholar
- Liu, F., Li, H., Ren, C., Bo, X. & Shu, W. PEDLA: predicting enhancers with a deep learning-based algorithmic framework. Sci. Rep.6, 28517 (2016). CASPubMed CentralPubMedGoogle Scholar
- Li, Y., Shi, W. & Wasserman, W. W. Genome-wide prediction of cis-regulatory regions using supervised deep learning methods. BMC Bioinformatics19, 202 (2018). PubMed CentralPubMedGoogle Scholar
- Johnson, D. S., Mortazavi, A., Myers, R. M. & Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science316, 1497–1502 (2007). CASPubMedGoogle Scholar
- Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell129, 823–837 (2007). CASPubMedGoogle Scholar
- Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods4, 651–657 (2007). CASPubMedGoogle Scholar
- Park, P. J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet.10, 669–680 (2009). CASPubMed CentralPubMedGoogle Scholar
- Weirauch, M. T. et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol.31, 126 (2013). CASPubMed CentralPubMedGoogle Scholar
- Lee, D., Karchin, R. & Beer, M. A. Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res.21, 2167–2180 (2011). CASPubMed CentralPubMedGoogle Scholar
- Ghandi, M., Lee, D., Mohammad-Noori, M. & Beer, M. A. Enhanced regulatory sequence prediction using gapped k-mer features. PLOS Comput. Biol.10, e1003711 (2014). PubMed CentralPubMedGoogle Scholar
- Stormo, G. D., Schneider, T. D., Gold, L. & Ehrenfeucht, A. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res.10, 2997–3011 (1982). CASPubMed CentralPubMedGoogle Scholar
- Stormo, G. D. DNA binding sites: representation and discovery. Bioinformatics16, 16–23 (2000). CASPubMedGoogle Scholar
- D’haeseleer, P. What are DNA sequence motifs? Nat. Biotechnol.24, 423–425 (2006). PubMedGoogle Scholar
- Kelley, D. R., Snoek, J. & Rinn, J. L. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res.26, 990–999 (2016). This paper describes the application of a deep CNN to predict chromatin accessibility in 164 cell types from DNA sequence. CASPubMed CentralPubMedGoogle Scholar
- Wang, M., Tai, C., E, W. & Wei, L. DeFine: deep convolutional neural networks accurately quantify intensities of transcription factor-DNA binding and facilitate evaluation of functional non-coding variants. Nucleic Acids Res.46, e69 (2018). PubMed CentralPubMedGoogle Scholar
- Kelley, D. R. et al. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res.28, 739–750 (2018). In this paper, a deep CNN was trained to predict more than 4,000 genomic measurements including gene expression as measured by cap analysis of gene expression (CAGE) for every 150 bp in the genome using a receptive field of 32 kb. CASPubMed CentralPubMedGoogle Scholar
- Schreiber, J., Libbrecht, M., Bilmes, J. & Noble, W. Nucleotide sequence and DNaseI sensitivity are predictive of 3D chromatin architecture. Preprint at bioRxivhttps://doi.org/10.1101/103614 (2018). ArticleGoogle Scholar
- Zeng, H. & Gifford, D. K. Predicting the impact of non-coding variants on DNA methylation. Nucleic Acids Res.45, e99 (2017). PubMed CentralPubMedGoogle Scholar
- Angermueller, C., Lee, H. J., Reik, W. & Stegle, O. DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol.18, 67 (2017). PubMed CentralPubMedGoogle Scholar
- Zhou, J. et al. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat. Genet.50, 1171–1179 (2018). In this paper, two models, a deep CNN and a linear model, are stacked to predict tissue-specific gene expression from DNA sequence, which demonstrates the utility of this approach in non-coding variant effect prediction. CASPubMed CentralPubMedGoogle Scholar
- Cuperus, J. T. et al. Deep learning of the regulatory grammar of yeast 5’ untranslated regions from 500,000 random sequences. Genome Res.27, 2015–2024 (2017). CASPubMed CentralPubMedGoogle Scholar
- Pan, X. & Shen, H.-B. RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach. BMC Bioinformatics18, 136 (2017). PubMed CentralPubMedGoogle Scholar
- Avsec, Ž., Barekatain, M., Cheng, J. & Gagneur, J. Modeling positional effects of regulatory sequences with spline transformations increases prediction accuracy of deep neural networks. Bioinformatics34, 1261–1269 (2018). CASPubMedGoogle Scholar
- Budach, S. & Marsico, A. pysster: classification of biological sequences by learning sequence and structure motifs with convolutional neural networks. Bioinformatics34, 3035–3037 (2018). CASPubMed CentralPubMedGoogle Scholar
- Cheng, S. et al. MiRTDL: a deep learning approach for miRNA target prediction. IEEE/ACM Trans. Comput. Biol. Bioinform.13, 1161–1169 (2016). Google Scholar
- Kim, H. K. et al. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat. Biotechnol.36, 239–241 (2018). CASPubMedGoogle Scholar
- Koh, P. W., Pierson, E. & Kundaje, A. Denoising genome-wide histone ChIP-seq with convolutional neuralnetworks. Bioinformatics33, i225–i233 (2017). CASPubMed CentralPubMedGoogle Scholar
- Zhang, Y. et al. Enhancing Hi-C data resolution with deep convolutional neural network HiCPlus. Nat. Commun.9, 750 (2018). PubMed CentralPubMedGoogle Scholar
- Nielsen, A. A. K. & Voigt, C. A. Deep learning to predict the lab-of-origin of engineered DNA. Nat. Commun.9, 3135 (2018). PubMed CentralPubMedGoogle Scholar
- Luo, R., Sedlazeck, F. J., Lam, T.-W. & Schatz, M. Clairvoyante: a multi-task convolutional deep neural network for variant calling in single molecule sequencing. Preprint at bioRxivhttps://doi.org/10.1101/310458 (2018). ArticleGoogle Scholar
- Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol.36, 983–987 (2018). In this paper, a deep CNN is trained to call genetic variants from different DNA-sequencing technologies. CASPubMedGoogle Scholar
- Jaganathan, K. et al. Predicting splicing from primary sequence with deep learning. Cell176, 535–548 (2019). CASPubMedGoogle Scholar
- Elman, J. L. Finding structure in time. Cogn. Sci.14, 179–211 (1990). Google Scholar
- Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput.9, 1735–1780 (1997). CASPubMedGoogle Scholar
- Bai, S., Zico Kolter, J. & Koltun, V. An empirical evaluation of generic convolutional and recurrent networks for sequence modeling. Preprint at arXivhttps://arxiv.org/abs/1803.01271 (2018).
- Pan, X., Rijnbeek, P., Yan, J. & Shen, H.-B. Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks. BMC Genomics19, 511 (2018). PubMed CentralPubMedGoogle Scholar
- Quang, D. & Xie, X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res.44, e107 (2016). PubMed CentralPubMedGoogle Scholar
- Quang, D. & Xie, X. FactorNet: a deep learning framework for predicting cell type specific transcription factor binding from nucleotide-resolution sequential data. Preprint at bioRxivhttps://doi.org/10.1101/151274 (2017). ArticleGoogle Scholar
- Lee, B., Baek, J., Park, S. & Yoon, S. in Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics 434–442 (ACM, 2016).
- Park, S., Min, S., Choi, H. & Yoon, S. deepMiRGene: deep neural network based precursor microRNA prediction. Preprint at arXivhttps://arxiv.org/abs/1605.00017 (2016).
- Boža, V., Brejová, B. & Vinař;, T. DeepNano: deep recurrent neural networks for base calling in MinION nanopore reads. PLOS ONE12, e0178751 (2017). PubMed CentralPubMedGoogle Scholar
- Mikheyev, A. S. & Tin, M. M. Y. A first look at the Oxford Nanopore MinION sequencer. Mol. Ecol. Resour.14, 1097–1102 (2014). CASPubMedGoogle Scholar
- Barabási, A.-L., Gulbahce, N. & Loscalzo, J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet.12, 56–68 (2011). PubMed CentralPubMedGoogle Scholar
- Mitra, K., Carvunis, A.-R., Ramesh, S. K. & Ideker, T. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet.14, 719–732 (2013). CASPubMed CentralPubMedGoogle Scholar
- Scarselli, F., Gori, M., Tsoi, A. C., Hagenbuchner, M. & Monfardini, G. The graph neural network model. IEEE Trans. Neural Netw.20, 61–80 (2009). PubMedGoogle Scholar
- Defferrard, M., Bresson, X. & Vandergheynst, P. in Advances in Neural Information Processing Systems 29 (NIPS 2016) (eds Lee, D. D., Sugiyama, M., Luxburg, U. V., Guyon, I. & Garnett, R.) 3844–3852 (Curran Associates Inc., 2016).
- Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. Preprint at arXivhttps://arxiv.org/abs/1609.02907 (2016).
- Battaglia, P. W. et al. Relational inductive biases, deep learning, and graph networks. Preprint at arXivhttps://arxiv.org/abs/1806.01261 (2018).
- Hamilton, W. L., Ying, R. & Leskovec, J. Inductive representation learning on large graphs. Preprint at arXivhttps://arxiv.org/abs/1706.02216 (2017).
- Chen, J., Ma, T. & Xiao, C. FastGCN: fast learning with graph convolutional networks via importance sampling. Preprint at arXivhttps://arxiv.org/abs/1801.10247 (2018).
- Zitnik, M. & Leskovec, J. Predicting multicellular function through multi-layer tissue networks. Bioinformatics33, i190–i198 (2017). CASPubMed CentralPubMedGoogle Scholar
- Zitnik, M., Agrawal, M. & Leskovec, J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics34, i457–i466 (2018). CASPubMed CentralPubMedGoogle Scholar
- Duvenaud, D. K. et al. in Advances in Neural Information Processing Systems 28 (NIPS2015) (eds Cortes, C., Lawrence, N. D., Lee, D. D., Sugiyama, M. & Garnett, R.) 2224–2232 (Curran Associates Inc., 2015).
- Kearnes, S., McCloskey, K., Berndl, M., Pande, V. & Riley, P. Molecular graph convolutions: moving beyond fingerprints. J. Comput. Aided Mol. Des.30, 595–608 (2016). CASPubMed CentralPubMedGoogle Scholar
- Dutil, F., Cohen, J. P., Weiss, M., Derevyanko, G. & Bengio, Y. Towards gene expression convolutions using gene interaction graphs. Preprint at arXivhttps://arxiv.org/abs/1806.06975 (2018).
- Rhee, S., Seo, S. & Kim, S. in Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence 3527–3534 (IJCAI, 2018).
- Chen, Z., Badrinarayanan, V., Lee, C.-Y. & Rabinovich, A. GradNorm: gradient normalization for adaptive loss balancing in deep multitask networks. Preprint at arXivhttps://arxiv.org/abs/1711.02257 (2017).
- Sung, K. & Poggio, T. Example-based learning for view-based human face detection. IEEE Trans. Pattern Anal. Mach. Intell.20, 39–51 (1998). Google Scholar
- Felzenszwalb, P. F., Girshick, R. B., McAllester, D. & Ramanan, D. Object detection with discriminatively trained part-based models. IEEE Trans. Pattern Anal. Mach. Intell.32, 1627–1645 (2010). PubMedGoogle Scholar
- Guo, M., Haque, A., Huang, D.-A., Yeung, S. & Fei-Fei, L. in Computer Vision – ECCV 2018 (eds Ferrari, V., Hebert, M., Sminchisescu, C. & Weiss, Y.) Vol. 11220 282–299 (Springer International Publishing, 2018).
- Sundaram, L. et al. Predicting the clinical impact of human mutation with deep neural networks. Nat. Genet.50, 1161–1170 (2018). CASPubMed CentralPubMedGoogle Scholar
- Zitnik, M. et al. Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Inf. Fusion50, 71–91 (2018). PubMedPubMed CentralGoogle Scholar
- Yosinski, J., Clune, J., Bengio, Y. & Lipson, H. in Advances in Neural Information Processing Systems 27 (NIPS2014) (eds Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N. D. & Weinberger, K. Q.) 3320–3328 (Curran Associates Inc., 2014).
- Kornblith, S., Shlens, J. & Le, Q. V. Do better ImageNet models transfer better? Preprint at arXivhttps://arxiv.org/abs/1805.08974 (2018).
- Russakovsky, O. et al. ImageNet large scale visual recognition challenge. Preprint at arXivhttps://arxiv.org/abs/1409.0575 (2014).
- Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature542, 115–118 (2017). CASPubMedPubMed CentralGoogle Scholar
- Pawlowski, N., Caicedo, J. C., Singh, S., Carpenter, A. E. & Storkey, A. Automating morphological profiling with generic deep convolutional networks. Preprint at bioRxivhttps://doi.org/10.1101/085118 (2016). ArticleGoogle Scholar
- Zeng, T., Li, R., Mukkamala, R., Ye, J. & Ji, S. Deep convolutional neural networks for annotating gene expression patterns in the mouse brain. BMC Bioinformatics16, 147 (2015). PubMed CentralPubMedGoogle Scholar
- Zhang, W. et al. in IEEE Transactions on Big Data (IEEE, 2018).
- Adam, P. et al. Automatic differentiation in PyTorch. Presented at 31st Conference on Neural Information Processing Systems (NIPS 2017).
- Abadi, M. et al. Tensorflow: large-scale machine learning on heterogeneous distributed systems. Preprint at arXivhttps://arxiv.org/abs/1603.04467 (2016).
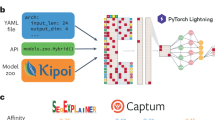
- Avsec, Z. et al. Kipoi: accelerating the community exchange and reuse of predictive models for genomics. Preprint at bioRxivhttps://doi.org/10.1101/375345 (2018).This paper describes a platform to exchange trained predictive models in genomics including deep neural networks. ArticleGoogle Scholar
- Breiman, L. Statistical modeling: the two cultures (with comments and a rejoinder by the author). Stat. Sci.16, 199–231 (2001). Google Scholar
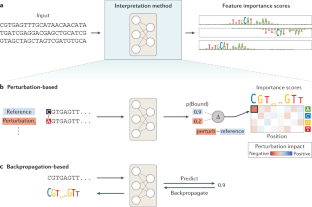
- Greenside, P., Shimko, T., Fordyce, P. & Kundaje, A. Discovering epistatic feature interactions from neural network models of regulatory DNA sequences. Bioinformatics34, i629–i637 (2018). CASPubMed CentralPubMedGoogle Scholar
- Zeiler, M. D. & Fergus, R. in Computer Vision – ECCV 2014 (eds Fleet, D., Pajdla, T., Schiele, B. & Tuytelaars, T.) Vol. 8689 818–833 (Springer International Publishing, 2014).
- Simonyan, K., Vedaldi, A. & Zisserman, A. Deep inside convolutional networks: visualising image classification models and saliency maps. Preprint at arXivhttps://arxiv.org/abs/1312.6034 (2013).
- Shrikumar, A., Greenside, P., Shcherbina, A. & Kundaje, A. Not just a black box: learning important features through propagating activation differences. Preprint at arXivhttps://arxiv.org/abs/1605.01713 (2016). This paper introduces DeepLIFT, a neural network interpretation method that highlights inputs most influential for the prediction.
- Sundararajan, M., Taly, A. & Yan, Q. Axiomatic attribution for deep networks. Preprint at arXivhttps://arxiv.org/abs/1703.01365 (2017).
- Lanchantin, J., Singh, R., Wang, B. & Qi, Y. Deep motif dashboard: visualizing and understanding genomic sequences using deep neural networks. Pac. Symp. Biocomput.22, 254–265 (2017). PubMed CentralPubMedGoogle Scholar
- Shrikumar, A. et al. TF-MoDISco v0.4.4.2-alpha: technical note. Preprint at arXivhttps://arxiv.org/abs/1811.00416v2 (2018).
- Ma, J. et al. Using deep learning to model the hierarchical structure and function of a cell. Nat. Methods15, 290–298 (2018). CASPubMed CentralPubMedGoogle Scholar
- Hinton, G. E. & Salakhutdinov, R. R. Reducing the dimensionality of data with neural networks. Science313, 504–507 (2006). CASPubMedGoogle Scholar
- Kramer, M. A. Nonlinear principal component analysis using autoassociative neural networks. AIChE J.37, 233–243 (1991). CASGoogle Scholar
- Vincent, P., Larochelle, H., Bengio, Y. & Manzagol, P.-A. in Proceedings of the 25th International Conference on Machine Learning 1096–1103 (ACM, 2008).
- Vincent, P., Larochelle, H., Lajoie, I., Bengio, Y. & Manzagol, P.-A. Stacked denoising autoencoders: learning useful representations in a deep network with a local denoising criterion. J. Mach. Learn. Res.11, 3371–3408 (2010). Google Scholar
- Jolliffe, I. in International Encyclopedia of Statistical Science (ed. Lovric, M.) 1094–1096 (Springer Berlin Heidelberg, 2011).
- Plaut, E. From principal subspaces to principal components with linear autoencoders. Preprint at arXivhttps://arxiv.org/abs/1804.10253 (2018).
- Kunin, D., Bloom, J. M., Goeva, A. & Seed, C. Loss landscapes of regularized linear autoencoders. Preprint at arXivhttps://arxiv.org/abs/1901.08168 (2019).
- Scholz, M., Kaplan, F., Guy, C. L., Kopka, J. & Selbig, J. Non-linear PCA: a missing data approach. Bioinformatics21, 3887–3895 (2005). CASPubMedGoogle Scholar
- Tan, J., Hammond, J. H., Hogan, D. A. & Greene, C. S. ADAGE-based integration of publicly available Pseudomonas aeruginosa gene expression data with denoising autoencoders illuminates microbe-host interactions. mSystems1, e00025–15 (2016). PubMed CentralPubMedGoogle Scholar
- Tan, J. et al. ADAGE signature analysis: differential expression analysis with data-defined gene sets. BMC Bioinformatics18, 512 (2017). PubMed CentralPubMedGoogle Scholar
- Tan, J. et al. Unsupervised extraction of stable expression signatures from public compendia with an ensemble of neural networks. Cell Syst.5, 63–71 (2017). CASPubMed CentralPubMedGoogle Scholar
- Brechtmann, F. et al. OUTRIDER: a statistical method for detecting aberrantly expressed genes in RNA sequencing data. Am. J. Hum. Genet.103, 907–917 (2018). CASPubMed CentralPubMedGoogle Scholar
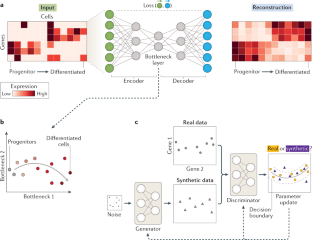
- Ding, J., Condon, A. & Shah, S. P. Interpretable dimensionality reduction of single cell transcriptome data with deep generative models. Nat. Commun.9, 2002 (2018). PubMed CentralPubMedGoogle Scholar
- Cho, H., Berger, B. & Peng, J. Generalizable and scalable visualization of single-cell data using neural networks. Cell Syst.7, 185–191 (2018). CASPubMed CentralPubMedGoogle Scholar
- Deng, Y., Bao, F., Dai, Q., Wu, L. & Altschuler, S. Massive single-cell RNA-seq analysis and imputation via deep learning. Preprint at bioRxivhttps://doi.org/10.1101/315556 (2018). ArticleGoogle Scholar
- Talwar, D., Mongia, A., Sengupta, D. & Majumdar, A. AutoImpute: autoencoder based imputation of single-cell RNA-seq data. Sci. Rep.8, 16329 (2018). PubMed CentralPubMedGoogle Scholar
- Amodio, M. et al. Exploring single-cell data with deep multitasking neural networks. Preprint at bioRxivhttps://doi.org/10.1101/237065 (2019). ArticleGoogle Scholar
- Eraslan, G., Simon, L. M., Mircea, M., Mueller, N. S. & Theis, F. J. Single-cell RNA-seq denoising using a deep count autoencoder. Nat. Commun.10, 390 (2019). PubMed CentralPubMedGoogle Scholar
- Lin, C., Jain, S., Kim, H. & Bar-Joseph, Z. Using neural networks for reducing the dimensions of single-cell RNA-Seq data. Nucleic Acids Res.45, e156 (2017). PubMed CentralPubMedGoogle Scholar
- Kingma, D. P. & Welling, M. Auto-encoding variational bayes. Preprint at arXivhttps://arxiv.org/abs/1312.6114 (2013).
- Goodfellow, I. et al. in Advances in Neural Information Processing Systems 27 (NIPS2014) (eds Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N. D. & Weinberger, K. Q.) 2672–2680 (Curran Associates Inc., 2014).
- Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods15, 1053–1058 (2018). CASPubMed CentralPubMedGoogle Scholar
- Way, G. P. & Greene, C. S. in Biocomputing 2018: Proceedings of the Pacific Symposium (eds Altman, R. B. et al.) 80–91 (World Scientific, 2018).
- Grønbech, C. H. et al. scVAE: variational auto-encoders for single-cell gene expression data. Preprint at bioRxivhttps://doi.org/10.1101/318295 (2018).
- Wang, D. & Gu, J. VASC: dimension reduction and visualization of single-cell RNA-seq data by deep variational autoencoder. Genomics Proteomics Bioinformatics16, 320–331 (2018). PubMed CentralPubMedGoogle Scholar
- Lotfollahi, M., Alexander Wolf, F. & Theis, F. J. Generative modeling and latent space arithmetics predict single-cell perturbation response across cell types, studies and species. Preprint at bioRxivhttps://doi.org/10.1101/478503 (2018). ArticleGoogle Scholar
- Hu, Q. & Greene, C. S. Parameter tuning is a key part of dimensionality reduction via deep variational autoencoders for single cell RNA transcriptomics. Preprint at bioRxivhttps://doi.org/10.1101/385534 (2018). ArticleGoogle Scholar
- Gupta, A. & Zou, J. Feedback GAN (FBGAN) for DNA: a novel feedback-loop architecture for optimizing protein functions. Preprint at arXivhttps://arxiv.org/abs/1804.01694 (2018).
- Killoran, N., Lee, L. J., Delong, A., Duvenaud, D. & Frey, B. J. Generating and designing DNA with deep generative models. Preprint at arXivhttps://arxiv.org/abs/1712.06148 (2017).
- Ghahramani, A., Watt, F. M. & Luscombe, N. M. Generative adversarial networks simulate gene expression and predict perturbations in single cells. Preprint at bioRxivhttps://doi.org/10.1101/262501 (2018). ArticleGoogle Scholar
- Amodio, M. & Krishnaswamy, S. MAGAN: aligning biological manifolds. Preprint at arXivhttps://arxiv.org/abs/1803.00385 (2018).
- Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science337, 1190–1195 (2012). CASPubMed CentralPubMedGoogle Scholar
- Cheng, J. et al. MMSplice: modular modeling improves the predictions of genetic variant effects on splicing. Genome Biol.20, 48 (2019). PubMed CentralPubMedGoogle Scholar
- van der Maaten, L. in Proceedings of the Twelfth International Conference on Artificial Intelligence and Statistics (eds van Dyk, D. & Welling, M.) Vol. 5 384–391 (PMLR, 2009).
- Angerer, P. et al. Single cells make big data: new challenges and opportunities in transcriptomics. Curr. Opin. Syst. Biol.4, 85–91 (2017). Google Scholar
- Shaham, U. et al. Removal of batch effects using distribution-matching residual networks. Bioinformatics33, 2539–2546 (2017). CASPubMed CentralPubMedGoogle Scholar
- Regev, A. et al. The human cell atlas. eLife6, e27041 (2017). PubMed CentralPubMedGoogle Scholar
- Fleming, N. How artificial intelligence is changing drug discovery. Nature557, S55–S57 (2018). CASPubMedGoogle Scholar
- Kalinin, A. A. et al. Deep learning in pharmacogenomics: from gene regulation to patient stratification. Pharmacogenomics19, 629–650 (2018). CASPubMed CentralPubMedGoogle Scholar
- AlQuraishi, M. End-to-end differentiable learning of protein structure. Preprint at bioRxivhttps://doi.org/10.1101/265231 (2018). ArticleGoogle Scholar
- Nawy, T. Spatial transcriptomics. Nat. Methods15, 30 (2018). CASGoogle Scholar
- Eulenberg, P. et al. Reconstructing cell cycle and disease progression using deep learning. Nat. Commun.8, 463 (2017). PubMed CentralPubMedGoogle Scholar
- KoneČný, J., McMahan, H. B., Ramage, D. & Richtárik, P. Federated optimization: distributed machine learning for on-device intelligence. Preprint at arXivhttps://arxiv.org/abs/1610.02527 (2016).
- Beaulieu-Jones, B. K. et al. Privacy-preserving generative deep neural networks support clinical data sharing. Preprint at bioRxivhttps://doi.org/10.1101/159756 (2018).
- Lever, J., Krzywinski, M. & Altman, N. Classification evaluation. Nat. Methods13, 603 (2016). CASGoogle Scholar
- Tieleman, T. & Hinton, G. Lecture 6.5 - RMSProp, COURSERA: neural networks for machine learning (2012).
- Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Preprint at arXivhttps://arxiv.org/abs/1412.6980 (2014).
- Schmidhuber, J. Deep learning in neural networks: an overview. Neural Netw.61, 85–117 (2015). PubMedGoogle Scholar
- LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature521, 436–444 (2015). CASPubMedGoogle Scholar
- Bottou, L. in Proceedings of Neuro-Nımes ‘91 12 (EC2, 1991).
- Bengio, Y. Practical recommendations for gradient-based training of deep architectures. Preprint at arXivhttps://arxiv.org/abs/1206.5533 (2012).
- Bergstra, J. & Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res.13, 281–305 (2012). Google Scholar
- Bergstra, J., Yamins, D. & Cox, D. in Proceedings of the 30th International Conference on Machine Learning Vol. 28 115–123 (JMLR W&CP, 2013).
- Shahriari, B., Swersky, K., Wang, Z., Adams, R. P. & de Freitas, N. Taking the human out of the loop: a review of bayesian optimization. Proc. IEEE104, 148–175 (2016). Google Scholar
- Li, L., Jamieson, K., DeSalvo, G., Rostamizadeh, A. & Talwalkar, A. Hyperband: a novel bandit-based approach to hyperparameter optimization. J. Mach. Learn. Res.18, 6765–6816 (2017). Google Scholar
- Elsken, T., Metzen, J. H. & Hutter, F. Neural architecture search: a survey. Preprint at arXivhttps://arxiv.org/abs/1808.05377 (2018).
Acknowledgements
Ž.A. was supported by the German Bundesministerium für Bildung und Forschung (BMBF) through the project MechML (01IS18053F). The authors acknowledge M. Heinig and A. Raue for valuable feedback.
Reviewer information
Nature Reviews Genetics thanks C. Greene and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
- These authors contributed equally: Gökcen Eraslan, Žiga Avsec.
Authors and Affiliations
- Institute of Computational Biology, Helmholtz Zentrum München, Neuherberg, Germany Gökcen Eraslan & Fabian J. Theis
- School of Life Sciences Weihenstephan, Technical University of Munich, Freising, Germany Gökcen Eraslan & Fabian J. Theis
- Department of Informatics, Technical University of Munich, Garching, Germany Žiga Avsec & Julien Gagneur
- Department of Mathematics, Technical University of Munich, Garching, Germany Fabian J. Theis
- Gökcen Eraslan










